Publications - Prof. Dr. R. Santoro
Full list of publications
Featured publications
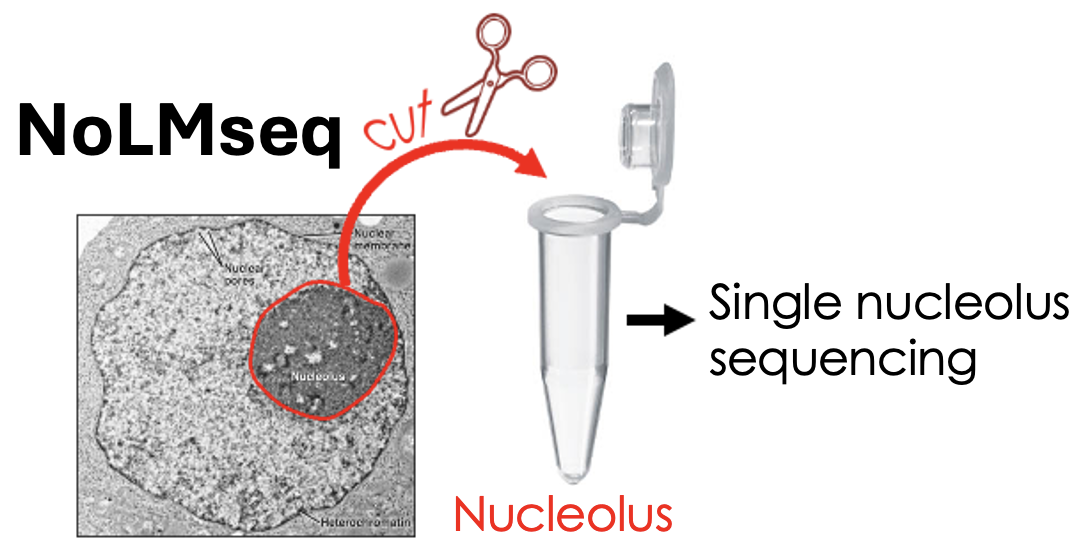
Walavalkar K, Gupta S, Kresoja-Rakic J, Raingeval M, Mungo C, Santoro R
Single-cell dynamics of genome-nucleolus interactions captured by nucleolar laser microdissection (NoLMseq)
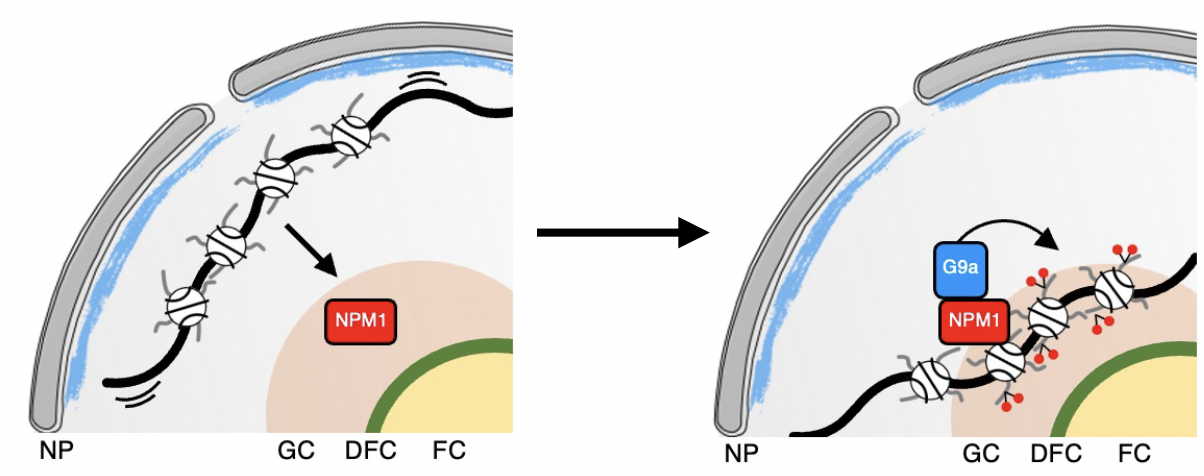
Gupta S, Bersaglieri C, Bär D, Raingeval M, Schaab L, Santoro R
NPM1 mediates genome-nucleolus interactions and the establishment of their repressive chromatin states
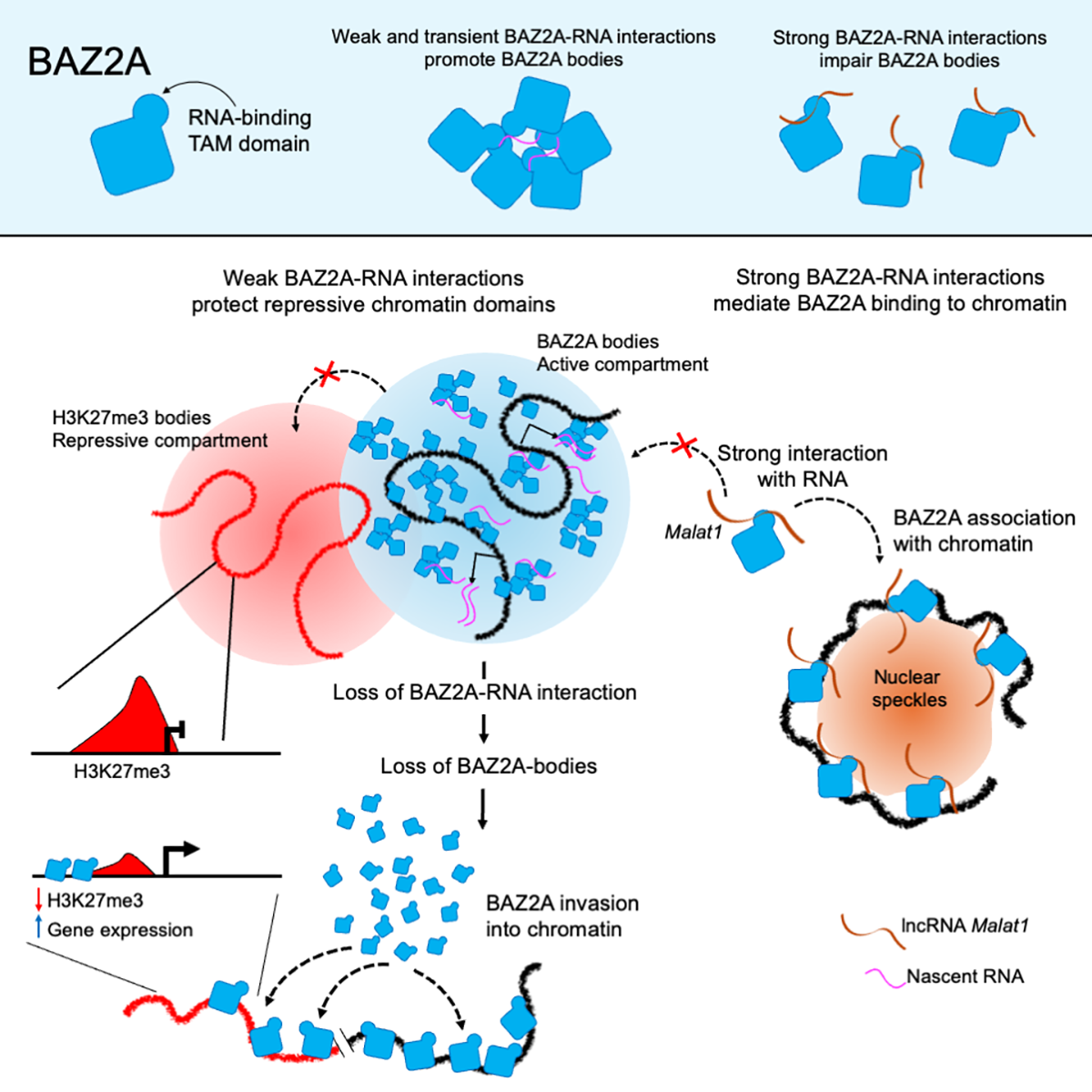
Lerra L, Panatta M, Bär D, Zanini I, Yihong Tan J, Pisano A, Mungo C, Baroux C, Govind Panse V, Marques AC, Santoro R
An RNA-dependent and phase-separated active subnuclear compartment safeguards repressive chromatin domains
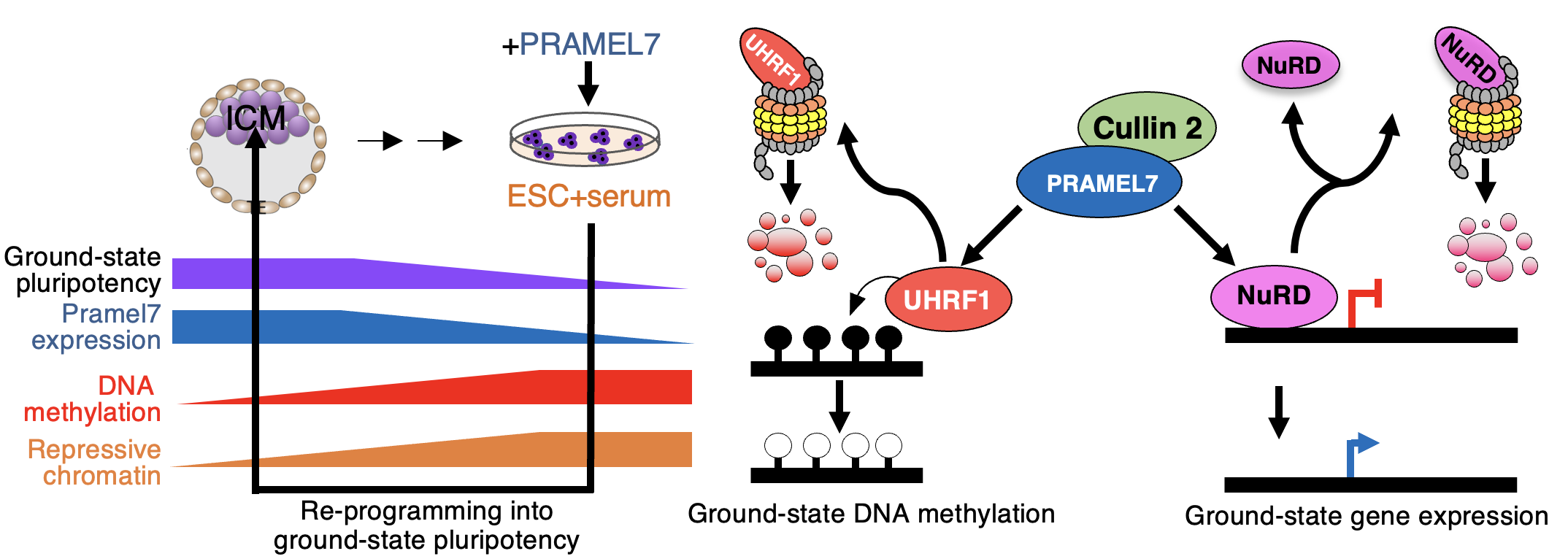
Rupasinghe M, Bersaglieri C, Leslie Pedrioli DM, Pedrioli PGA, Panatta M, Hottiger MO, Cinelli P, Santoro R
PRAMEL7 and CUL2 decrease NuRD stability to establish ground-state pluripotency
EMBO Reports (2024) 25(3):1453-1468
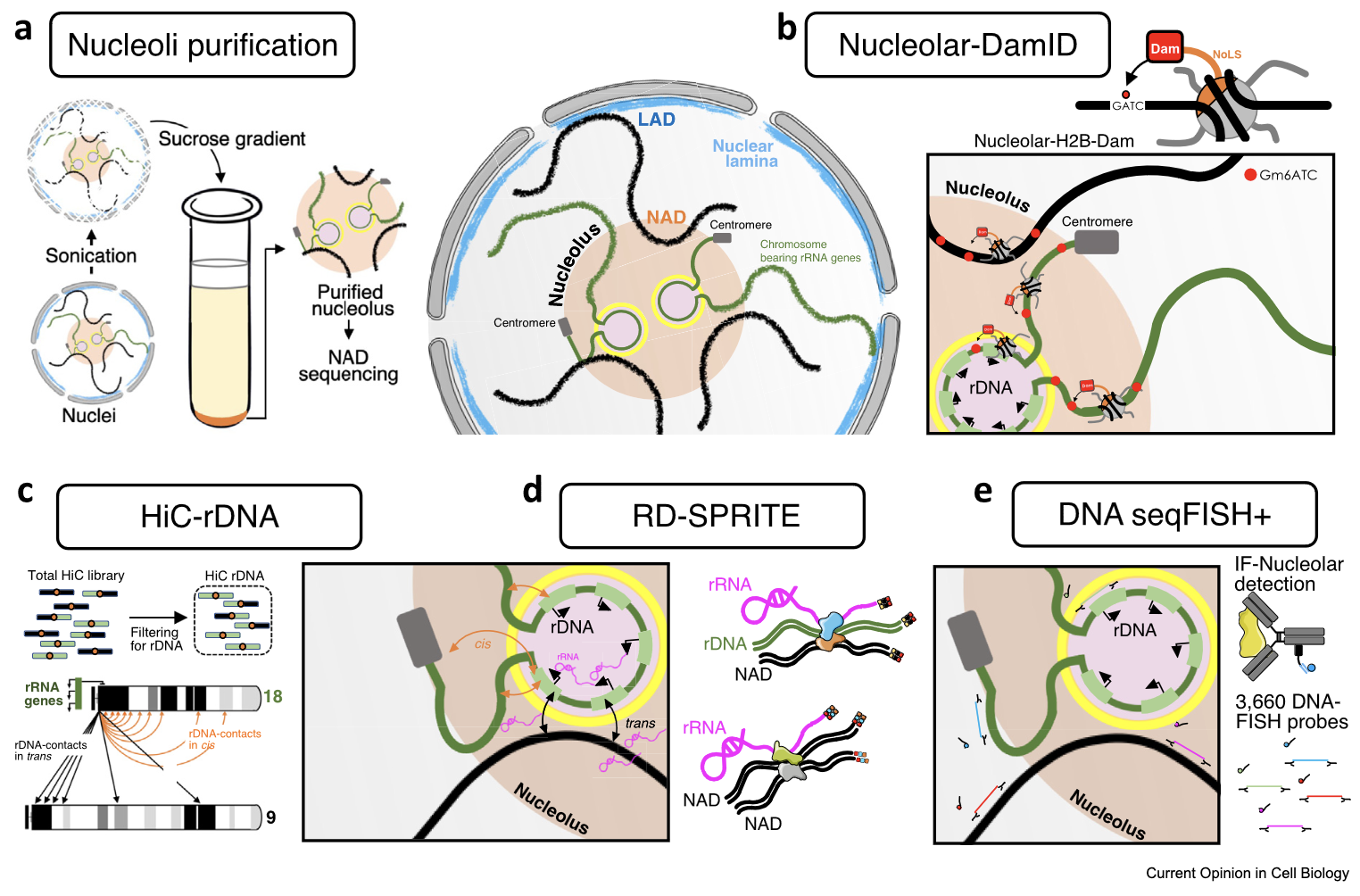
Bersaglieri C, Santoro R
Methods for mapping 3D-chromosome architecture around nucleoli
Curr Opin Cell Biol (2023) 81:102171
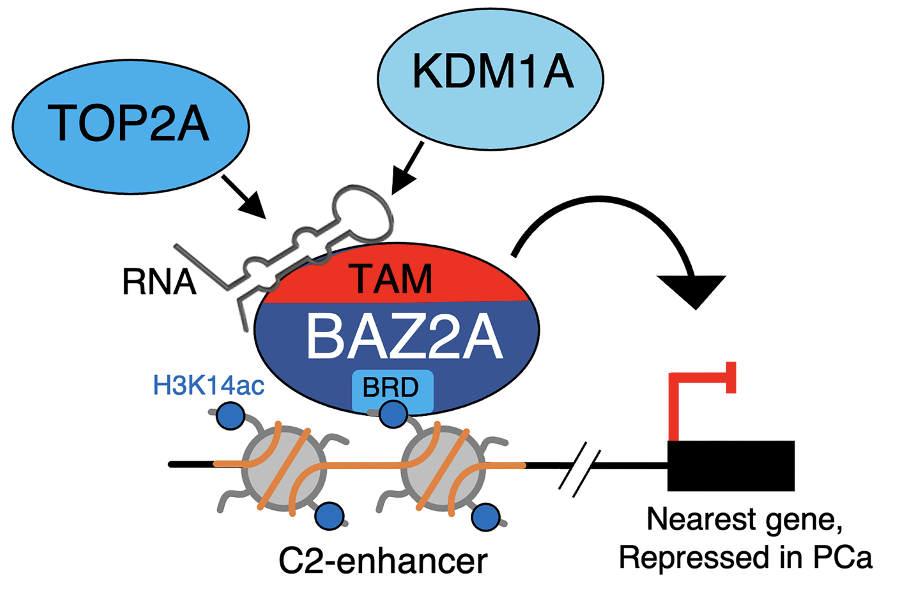
Roganowicz M, Bär D, Bersaglieri C, Aprigliano R, Santoro R
BAZ2A-RNA mediated association with TOP2A and KDM1A represses genes implicated in prostate cancer
Life Science Alliance (2023) 6(7):e202301950
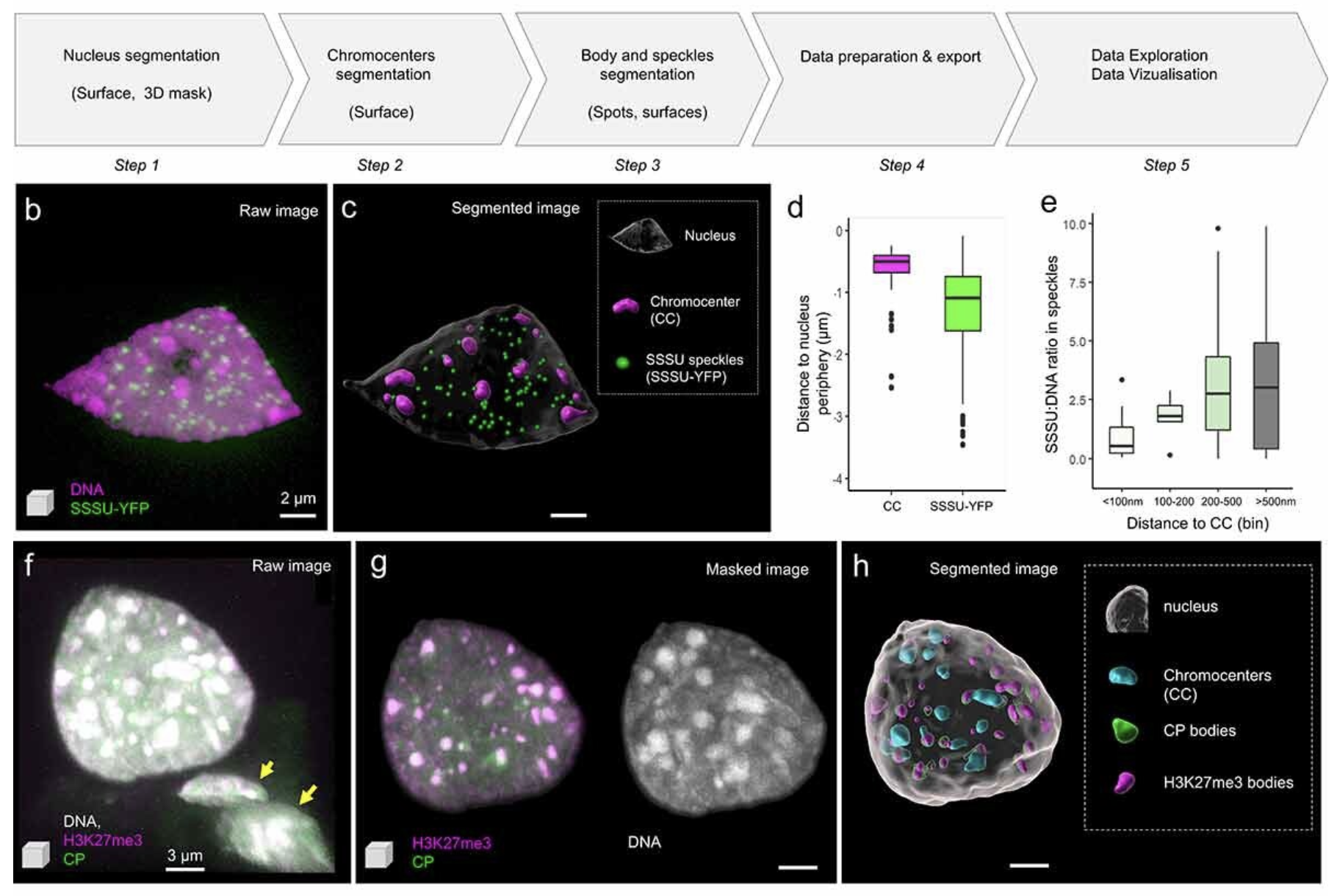
Randall RS, Jourdain C, Nowicka A, Kaduchová K, Kubová M, Ayoub MA, Schubert V, Tatout C, Colas I, Kalyanikrishna, Desset S, Mermet S, Boulaflous-Stevens A, Kubalová I, Mandáková T, Heckmann S, Lysak MA, Panatta M, Santoro R, Schubert D, Pecinka A, Routh D, Baroux C
Image analysis workflows to reveal the spatial organization of cell nuclei and chromosomes
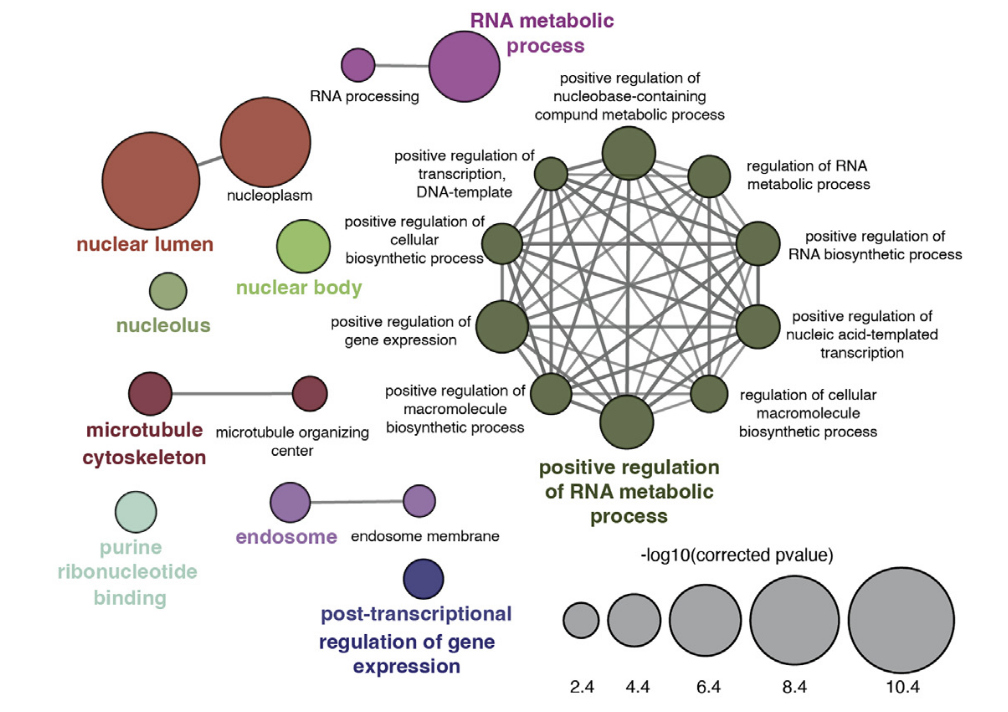
Müller M, Schaefer M, Fäh T, Spies D, Hermes V, Ngondo RP, Peña-Hernández R, Santoro R, Ciaudo C
Argonaute proteins regulate a specific network of genes through KLF4 in mouse embryonic stem cells
Stem Cell Reports (2022) 17(5):1070-1080

Bersaglieri C, Kresoja-Rakic J, Gupta S, Bär D, Kuzyakiv R, Panatta M, Santoro R
Genome-wide maps of nucleolus interactions reveal distinct layers of repressive chromatin domains

Raffaella Santoro
Looking for a job in a dynamic and collaborative working place? LncRNAs are recruiting!

Peña-Hernández R, Aprigliano R, Frommel S, Pietrzak K, Steiger S, Roganowicz M, Lerra L, Bizzarro J, Santoro R
BAZ2A-mediated repression via H3K14ac-marked enhancers promotes prostate cancer stem cells

Diener J, Baggiolini A, Pernebrink M, Dalcher D, Lerra L, Cheng PF, Varum S, Häusel J, Stierli S, Treier M, Studer L, Basler K, Levesque MP, Dummer R, Santoro R, Cantù C, Sommer L
Epigenetic control of melanoma cell invasiveness by the stem cell factor SALL4


Dalcher D, Yihong Tan J, Bersaglieri C, Peña-Hernández R, Vollenweider E, Zeyen S, Schmid MW, Bianchi V, Butz S, Roganowicz M, Kuzyakiv R, Baubec T, Marques AC, Santoro R
BAZ2A safeguards genome architecture of ground-state pluripotent stem cells
The EMBO Journal (2020) e105606

Kresoja-Rakic J, Santoro R
Nucleolus and rRNA Gene Chromatin in Early Embryo Development
Trends Genet. 2019 Jul 35(11): 868-879

Bersaglieri C, Santoro R
Genome Organization in and around the Nucleolus
Cells 2019 Jun 12;8(6). pii: E579

Gupta S, Santoro R
Regulation and Roles of the Nucleolus in Embryonic Stem Cells: From Ribosome Biogenesis to Genome Organization
Stem Cell Reports. 2020 Sep 8:S2213-6711(20)30343-X

Pietrzak K, Kuzyakiv R, Simon R, Bolis M, Bär D, Aprigliano R, Theurillat JP, Sauter G, Santoro R
TIP5 primes prostate luminal cells for the oncogenic transformation mediated by PTEN-loss
PNAS February 18, 2020 117 (7) 3637-3647

Wyck S, Herrera C, Requena CE, Bittner L, Hajkova P, Bollwein H, Santoro R
Oxidative stress in sperm affects the epigenetic reprogramming in early embryonic development
Epigenetics Chromatin. 2018 Oct 17;11(1):60

Zingg D, Debbache J, Peña-Hernández R, Antunes AT, Schaefer SM, Cheng PF, Zimmerli D, Haeusel J, Calçada RR, Tuncer E, Zhang Y, Bossart R, Wong KK, Basler K, Dummer R, Santoro R, Levesque MP, Sommer L
EZH2-Mediated Primary Cilium Deconstruction Drives Metastatic Melanoma Formation
Cancer Cell. 2018 Jul 9;34(1):69-84 e 14.
Graf U, Casanova E, Wyck S, Dalcher D, Gatti M, Vollenweider E, Okoniewski M, Weber FA, Patel SS, Schmid M, Li J, Sharif J, Wanner GA, Koseki H, Wong J, Pelczar P, Penengo L, Santoro R*, Cinelli P*
*Senior and corresponding authors
Pramel7 mediates ground state pluripotency through proteasomal-epigenetic combined pathways
Nature Cell Biology (2017) 19, 763-773UZH press release
Leone S, Bär D, Slabber CF, Dalcher D, Santoro R
The RNA helicase DHX9 establishes nucleolar heterochromatin and this activity is required for embryonic stem cell differentiation
EMBO Reports 2017 Jun; 18:1248-1262
Leone S, Santoro R
Challenges in the analysis of long noncoding RNA functionality
FEBS Letter 2016 Aug;590(15):2342-2353
Böhm M, Wachte M, Marques J, Streiff N, Nanni P, Mamchaoui K, Santoro R, Schäfer BW
Helicase CHD4 functions as epigenetic co-regulator of PAX3-FOXO1 activity in alveolar rhabdomyosarcoma
Journal of Clinical Investigation 2016 Nov 126(11): 4237-4249
Kresoja-Rakic J, Kapaklikaya E, Ziltener G, Dalcher D, Santoro R, Christensen BC, Johnson KC, Schwaller B, Weder W, Stahel RA, Felley-Bosco E
Identification of cis- and trans-acting elements regulating calretinin expression in mesothelioma cells

Gu L, Frommel SC, Oakes CC, Simon R, Grupp K, Gerig CY, Bär D, Robinson MD, Baer C, Weiss M, Gu Z, Schapira M, Kuner R, Sültmann H, Provenzano M; ICGC Project on Early Onset Prostate Cancer, Yaspo ML, Brors B, Korbel J, Schlomm T, Sauter G, Eils R, Plass C, Santoro R
BAZ2A (TIP5) is involved in epigenetic alterations in prostate cancer and its overexpression predicts disease recurrence
Nat Genet. 2015 Jan;47(1):22-30
Cheng PF, Shakhova O, Widmer DS, Eichhoff OM, Zingg D, Frommel SC, Belloni B, Raaijmakers MI, Goldinger SM, Santoro R, Hemmi S, Sommer L, Dummer R, Levesque MP
Methylation-dependent SOX9 expression mediates invasion in human melanoma cells and is a negative prognostic factor in advanced melanoma
Genome Biol. 2015 Feb 22;16:42
Shakhova O, Cheng P, Mishra PJ, Zingg D, Schaefer SM, Debbache J, Häusel J, Matter C, Guo T, Davis S, Meltzer P, Mihic-Probst D, Moch H, Wegner M, Merlino G, Levesque MP, Dummer R, Santoro R, Cinelli P, Sommer L
Antagonistic cross-regulation between Sox9 and Sox10 controls an anti-tumorigenic program in melanoma
PLoS Genet. 2015 Jan 28;11(1):e1004877
Zingg D, Debbache J, Schaefer SM, Tuncer E, Frommel SC, Cheng P, Arenas-Ramirez N, Haeusel J, Zhang Y, Bonalli M, McCabe MT, Creasy CL, Levesque MP, Boyman O, Santoro R, Shakhova O, Dummer R, Sommer L
The epigenetic modifier EZH2 controls melanoma growth and metastasis through silencing of distinct tumour suppressors
Nat Commun. 2015 Jan 22;6:6051

Savić N, Bär D, Leone S, Frommel SC, Weber FA, Vollenweider E, Ferrari E, Ziegler U, Kaech A, Shakhova O, Cinelli P, Santoro R
lncRNA Maturation to Initiate Heterochromatin Formation in the Nucleolus Is Required for Exit from Pluripotency in ESCs
Cell Stem Cell. 2014 Dec 4;15(6):720-34
Highlighted in The Nucleolus Gets the Silent Treatment. Cell Stem Cell. 2014 Dec 4;15(6):675-6
Bachmann SB, Frommel SC, Camicia R, Winkler HC, Santoro R, Hassa PO
DTX3L and ARTD9 inhibit IRF1 expression and mediate in cooperation with ARTD8 survival and proliferation of metastatic prostate cancer cells
Mol Cancer. 2014 May 27;13:125
Léger K, Bär D, Savić N, Santoro R, Hottiger MO
ARTD2 activity is stimulated by RNA
Nucleic Acids Res. 2014 Apr;42(8):5072-82
Santoro R
Analysis of chromatin composition of repetitive sequences: the ChIP-Chop assay
Methods Mol Biol. 2014;1094:319-28
Dantzer F, Santoro R
The expanding role of PARPs in the establishment and maintenance of heterochromatin
FEBS J. 2013 Jun 3. doi: 10.1111/febs.12368

Guetg C, Santoro R
Formation of nuclear heterochromatin: the nucleolar point of view
Epigenetics. 2012 Aug 1;7(8). ahead of print
Guetg C, Santoro R
Noncoding RNAs link PARP1 to heterochromatin
Cell Cycle. 2012 Jun 15;11(12):2217-8. Epub 2012 Jun 15.

Guetg C, Scheifele F, Rosenthal F, Hottiger MO, Santoro R
Inheritance of silent rDNA chromatin is mediated by PARP1 via non-coding RNA.
Molecular Cell 45, 790-800, 2012
Commented in Preview “PARP1 Parylation Promotes Silent Locus Transmission in the Nucleolus: The Suspicion Confirmed” Isabelle et al. Molecular Cell 45, 706-707, 2012
Erener E, Petrilli V, Kassner I, Minotti R, Castillo R, Santoro R, Hassa PO, Tschopp J, Hottiger MO
Inflammasome-activated Caspase 7 cleaves PARP1 to enhance the expression of a subset of NF-kb target genes.
Molecular Cell 10.1016/j.molcel.2012.02.016, 2012
Santoro R
The epigenetics of the nucleolus: structure and function of active and silent ribosomal RNA genes.
In “The Nucleolus”, Protein Reviews series, Springer, ed M.Olson. 15, 57-82, 2012
Osorio G, Varela I, Lara E, Puente XS, Espada J, Santoro R, Freije JMP, Fraga MF, López-Otín C
Nuclear envelope alterations generate an aging-like epigenetic pattern in mice deficient in Zmpste24 metalloprotease.
Guetg C, Lienemann P, Sirri V, Grummt I, Hernandez-Verdu D, Hottiger MO, Fussenegger M, Santoro R
The NoRC complex mediates heterochromatin and stability of silent rRNA genes and centromeric repeats
EMBO J. 29(13): 2135-2146, 2010
Santoro R, Schmitz KM, Sandoval J, Grummt I
Intergenic transcripts originating from a subclass of ribosomal DNA repeats silence ribosomal RNA genes in trans.
Santoro R*, Lienemann P, Fussenegger M
Epigenetic engineering of ribosomal RNA genes enhances protein production.
Espada J, Ballestar E, Santoro R, Fraga MF, Villar-Garea A, Nemeth A, Lopez-Serra L, Ropero S, Aranda A, Orozco H, Moreno V, Juarranz A, Stockert JC, Längst G, Grummt I, Bickmore W, Esteller M
Epigenetic disruption of ribosomal rRNA genes and nucleolar architecture in DNA methyltransferase (Dnmt1) deficient cells.
Nucl. Acid Research 35, 2191-2198, 2007
Mayer C, Schmitz K, Li J, Grummt I, Santoro R
Intergenic transcripts regulate the epigenetic state of rRNA genes.
Santoro R*, Grummt I
Epigenetic mechanisms of rRNA silencing: Temporal order of NoRC recruitment, histone modifications, chromatin remodeling and DNA methylation.
Mol. Cell. Biol. 25, 2539-2546, 2005
Li J, Santoro R, Grummt I
The chromatin remodeling complex NoRC controls replication timing of rRNA genes.
Santoro R*, De Lucia F
Many players, one goal: how chromatin states are inherited during cell division.
Biochem. Cell Biol. 83, 332-343, 2005
Santoro R, Li J, Grummt I
The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription.
Nature Genetics 32, 393-96, 2002
Commented in News and Views “Uniting the paths to gene silencing” Pikaard and Lawrence
Nature Genet. 32, 340-341
Zhou Y, Santoro R, Grummt I
The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription.
Santoro R, Grummt I
Molecular mechanisms mediating methylation dependent silencing of ribosomal gene transcription.
Strohner R, Nemeth A, Jansa P, Hoffmann-Rohrer U, Santoro R, Längst G, Grummt I
NoRC - a novel member of mammalian ISWI-containing chromatin remodeling machines.
Santoro R, Wolfl S, Saluz HP
UV-Laser induced protein/DNA crosslinking reveals sequence variations of DNA elements bound by c-Jun in vivo.
Biochem. Biophys. Res. Commun. 256, 68-74, 1999
Reale A, Marenzi S, Santoro R, D´Erme M, Zardo G, Strom R, Caiafa P
H1-H1 cross-linking efficiency dependes on genomic DNA methylation.
Biochem. Biophys. Res. Commun. 227, 768-774, 1996
Zardo G, Santoro R, D´Erme M, Reale A, Guidobaldi L, Caiafa P, Strom R
Specific inhibitory effect of H1e histone somatic variant on in vitro DNA methylation process.
Biochem. Biophys. Res. Commun. 220, 102-107, 1996
Santoro R, D´Erme M, Mastrantonio S, Reale A, Marenzi S, Zardo G, Caiafa P
Specific variants of H1 histone regulate CpG methylation in eukaryotic DNA.
Caiafa P, Reale A, Santoro R, D´Erme M, Marenzi S, Strom R
Does hypomethylation of linker DNA play a role in chromatin condensantion?
Santoro R, Di Matteo, Lavia P
Hydrazin to study DNA methylation, conformation and interaction with proteins.
In “DNA and Nucleoprotein Structure in vivo“, Springer-Verlag, ed. H.P. Saluz 1995
Santoro R, D´Erme M, Mastrantonio S, Reale A, Marenzi S, Saluz HP, Strom R, Caiafa P
Binding of H1e-c variants to CpG-rich DNA correlates with the inhibitory effect on enzymatic DNA methylation.
Biochem. Journal 305, 739-744, 1994
Santoro R, D´Erme M, Reale A, Marenzi S, Strom R, Caiafa P
Effect of H1 histone isoforms on the methylation of single- or double-stranded DNA.
Biochem. Biophys. Res. Commun. 190, 86-91, 1993
D´Erme M, Santoro R, Reale A, Marenzi S, Strom R, Caiafa P
Inhibition of CpG methylation in linker DNA by H1 histone.
Biochem. Biophys. Acta 1173, 209-216, 1993
Caiafa P, Reale A, D´Erme M, Allegra P, Santoro R, Strom R, Caiafa P
Histones and DNA methylation in mammalian chromatin. II presence of non-inhibitory tightly-bound histones.
Biochem. Biophys. Acta 1129, 43-48, 1991